2016
|
Radi M; Schneider R; Fallacara AL; Botta L; Crespan E; Tintori C; Maga G; Kissova M; Calgani A; Richters A; Musumeci F; Rauh D; Schenone S A cascade screening approach for the identification of Bcr-Abl myristate pocket binders active against wild type and T315I mutant. Journal Article In: Bioorganic & Medicinal Chemistry Letters, vol. 26, no 15, pp. 3436-3440, 2016. @article{%a1:%Y_304,
title = {A cascade screening approach for the identification of Bcr-Abl myristate pocket binders active against wild type and T315I mutant.},
author = {Radi M and Schneider R and Fallacara AL and Botta L and Crespan E and Tintori C and Maga G and Kissova M and Calgani A and Richters A and Musumeci F and Rauh D and Schenone S},
url = {http://www.sciencedirect.com/science/article/pii/S0960894X1630662X},
doi = {10.1016/j.bmcl.2016.06.051},
year = {2016},
date = {2016-02-17},
journal = {Bioorganic & Medicinal Chemistry Letters},
volume = {26},
number = {15},
pages = {3436-3440},
abstract = {The major clinical challenge in drug-resistant chronic myelogenous leukemia (CML) is currently represented by the Bcr-Abl T315I mutant, which is unresponsive to treatment with common first and second generation ATP-competitive tyrosine kinase inhibitors (TKIs). Allosteric inhibition of Bcr-Abl represent a new frontier in the fight against resistant leukemia and few candidates have been identified in the last few years. Among these, myristate pocket (MP) binders discovered by Novartis (e.g. GNF2/5) showed promising results, although they proved to be active against the T315I mutant only in combination with first and second generation ATP-competitive inhibitors. Here we used a cascade screening approach based on sequential fluorescence polarization (FP) screening, in silico docking/dynamics studies and kinetic-enzymatic studies to identify novel MP binders. A pyrazolo[3,4-d]pyrimidine derivative (6) has been identified as a promising allosteric inhibitor active on 32D leukemia cell lines (expressing Bcr-Abl WT and T315I) with no need of combination with any ATP-competitive inhibitor.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
The major clinical challenge in drug-resistant chronic myelogenous leukemia (CML) is currently represented by the Bcr-Abl T315I mutant, which is unresponsive to treatment with common first and second generation ATP-competitive tyrosine kinase inhibitors (TKIs). Allosteric inhibition of Bcr-Abl represent a new frontier in the fight against resistant leukemia and few candidates have been identified in the last few years. Among these, myristate pocket (MP) binders discovered by Novartis (e.g. GNF2/5) showed promising results, although they proved to be active against the T315I mutant only in combination with first and second generation ATP-competitive inhibitors. Here we used a cascade screening approach based on sequential fluorescence polarization (FP) screening, in silico docking/dynamics studies and kinetic-enzymatic studies to identify novel MP binders. A pyrazolo[3,4-d]pyrimidine derivative (6) has been identified as a promising allosteric inhibitor active on 32D leukemia cell lines (expressing Bcr-Abl WT and T315I) with no need of combination with any ATP-competitive inhibitor. |
Mentegari E; Kissova M; Bavagnoli L; Maga G; Crespan E DNA Polymerases lambda and beta: The Double-Edged Swords of DNA Repair. Journal Article In: Genes (Basel), vol. 7, no 9, pp. e57, 2016. @article{%a1:%Y_299,
title = {DNA Polymerases lambda and beta: The Double-Edged Swords of DNA Repair.},
author = {Mentegari E and Kissova M and Bavagnoli L and Maga G and Crespan E},
url = {http://www.mdpi.com/2073-4425/7/9/57},
doi = {10.3390/genes7090057},
year = {2016},
date = {2016-02-11},
journal = {Genes (Basel)},
volume = {7},
number = {9},
pages = {e57},
abstract = {DNA is constantly exposed to both endogenous and exogenous damages. More than 10,000 DNA modifications are induced every day in each cell's genome. Maintenance of the integrity of the genome is accomplished by several DNA repair systems. The core enzymes for these pathways are the DNA polymerases. Out of 17 DNA polymerases present in a mammalian cell, at least 13 are specifically devoted to DNA repair and are often acting in different pathways. DNA polymerases beta and lambda are involved in base excision repair of modified DNA bases and translesion synthesis past DNA lesions. Polymerase lambda also participates in non-homologous end joining of DNA double-strand breaks. However, recent data have revealed that, depending on their relative levels, the cell cycle phase, the ratio between deoxy- and ribo-nucleotide pools and the interaction with particular auxiliary proteins, the repair reactions carried out by these enzymes can be an important source of genetic instability, owing to repair mistakes. This review summarizes the most recent results on the ambivalent properties of these enzymes in limiting or promoting genetic instability in mammalian cells, as well as their potential use as targets for anticancer chemotherapy.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
DNA is constantly exposed to both endogenous and exogenous damages. More than 10,000 DNA modifications are induced every day in each cell's genome. Maintenance of the integrity of the genome is accomplished by several DNA repair systems. The core enzymes for these pathways are the DNA polymerases. Out of 17 DNA polymerases present in a mammalian cell, at least 13 are specifically devoted to DNA repair and are often acting in different pathways. DNA polymerases beta and lambda are involved in base excision repair of modified DNA bases and translesion synthesis past DNA lesions. Polymerase lambda also participates in non-homologous end joining of DNA double-strand breaks. However, recent data have revealed that, depending on their relative levels, the cell cycle phase, the ratio between deoxy- and ribo-nucleotide pools and the interaction with particular auxiliary proteins, the repair reactions carried out by these enzymes can be an important source of genetic instability, owing to repair mistakes. This review summarizes the most recent results on the ambivalent properties of these enzymes in limiting or promoting genetic instability in mammalian cells, as well as their potential use as targets for anticancer chemotherapy. |
Tintori C; Brai A; Dasso Lang MC; Deodato D; Greco AM; Bizzarri BM; Cascone L; Casian A; Zamperini C; Dreassi E; Crespan E; Maga G; Vanham G; Ceresola E; Canducci F; Arien KK; Botta M Development and in Vitro Evaluation of a Microbicide Gel Formulation for a Novel Non-Nucleoside Reverse Transcriptase Inhibitor Belonging to the N-Dihydroalkyloxybenzyloxopyrimidines (N-DABOs) Family. Journal Article In: Journal of medicinal chemistry, vol. 59, no 6, pp. 2747-2759, 2016. @article{%a1:%Y_314,
title = {Development and in Vitro Evaluation of a Microbicide Gel Formulation for a Novel Non-Nucleoside Reverse Transcriptase Inhibitor Belonging to the N-Dihydroalkyloxybenzyloxopyrimidines (N-DABOs) Family.},
author = {Tintori C and Brai A and Dasso Lang MC and Deodato D and Greco AM and Bizzarri BM and Cascone L and Casian A and Zamperini C and Dreassi E and Crespan E and Maga G and Vanham G and Ceresola E and Canducci F and Arien KK and Botta M},
url = {http://pubs.acs.org/doi/abs/10.1021/acs.jmedchem.5b01979},
doi = {10.1021/acs.jmedchem.5b01979},
year = {2016},
date = {2016-02-10},
journal = {Journal of medicinal chemistry},
volume = {59},
number = {6},
pages = {2747-2759},
abstract = {Preventing HIV transmission by the use of a vaginal microbicide is a topic of considerable interest in the fight against AIDS. Both a potent anti-HIV agent and an efficient formulation are required to develop a successful microbicide. In this regard, molecules able to inhibit the HIV replication before the integration of the viral DNA into the genetic material of the host cells, such as entry inhibitors or reverse transcriptase inhibitors (RTIs), are ideal candidates for prevention purpose. Among RTIs, S- and N-dihydroalkyloxybenzyloxopyrimidines (S-DABOs and N-DABOs) are interesting compounds active at nanomolar concentration against wild type of RT and with a very interesting activity against RT mutations. Herein, novel N-DABOs were synthesized and tested as anti-HIV agents. Furthermore, their mode of binding was studied by molecular modeling. At the same time, a vaginal microbicide gel formulation was developed and tested for one of the most promising candidates.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Preventing HIV transmission by the use of a vaginal microbicide is a topic of considerable interest in the fight against AIDS. Both a potent anti-HIV agent and an efficient formulation are required to develop a successful microbicide. In this regard, molecules able to inhibit the HIV replication before the integration of the viral DNA into the genetic material of the host cells, such as entry inhibitors or reverse transcriptase inhibitors (RTIs), are ideal candidates for prevention purpose. Among RTIs, S- and N-dihydroalkyloxybenzyloxopyrimidines (S-DABOs and N-DABOs) are interesting compounds active at nanomolar concentration against wild type of RT and with a very interesting activity against RT mutations. Herein, novel N-DABOs were synthesized and tested as anti-HIV agents. Furthermore, their mode of binding was studied by molecular modeling. At the same time, a vaginal microbicide gel formulation was developed and tested for one of the most promising candidates. |
Maga G Batteri Spazzini e Virus che curano: come le biotecnologie riscrivono la vita Book Zanichelli, Bologna, 2016, ISBN: 9788808920836. @book{CNRPRODOTTI368292,
title = {Batteri Spazzini e Virus che curano: come le biotecnologie riscrivono la vita},
author = {Maga G},
url = {http://www.zanichelli.it/ricerca/prodotti/batteri-spazzini-e-virus-che-curano},
isbn = {9788808920836},
year = {2016},
date = {2016-01-01},
publisher = {Zanichelli},
address = {Bologna},
keywords = {},
pubstate = {published},
tppubtype = {book}
}
|
2015
|
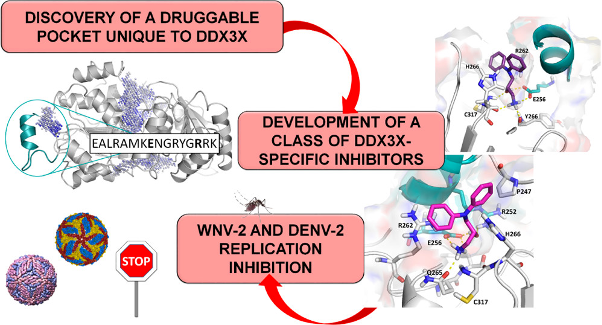
Fazi R; Tintori C; Brai A; Botta L; Selvaraj M; Garbelli A; Maga G; Botta M Homology Model-Based Virtual Screening for the Identification of Human Helicase DDX3 Inhibitors. Journal Article In: Journal of Chemical Information and Modeling, vol. 55, no 11, pp. 2443-2454, 2015. @article{%a1:%Y_370,
title = {Homology Model-Based Virtual Screening for the Identification of Human Helicase DDX3 Inhibitors.},
author = {Fazi R and Tintori C and Brai A and Botta L and Selvaraj M and Garbelli A and Maga G and Botta M},
url = {https://pubs.acs.org/doi/10.1021/acs.jcim.5b00419},
doi = {10.1021/acs.jcim.5b00419},
year = {2015},
date = {2015-11-23},
journal = {Journal of Chemical Information and Modeling},
volume = {55},
number = {11},
pages = {2443-2454},
abstract = {Targeting cellular cofactors instead of viral enzymes represents a new strategy to combat infectious diseases, which should help to overcome the problem of viral resistance. Recently, it has been revealed that the cellular ATPase/RNA helicase X-linked DEAD-box polypeptide 3 (DDX3) is an essential host factor for the replication of several viruses such as HIV, HCV, JEV, Dengue, and West Nile. Accordingly, a drug targeting DDX3 could theoretically inhibit all viruses that are dependent on this host factor. Herein, for the first time, a model of hDDX3 in its closed conformation, which binds the viral RNA was developed by using the homology module of Prime through the Maestro interface of Schrodinger. Next, a structure-based virtual screening protocol was applied to identify DDX3 small molecule inhibitors targeting the RNA binding pocket. As a result, an impressive hit rate of 40% was obtained with the identification of 10 active compounds out of the 25 tested small molecules. The best poses of the active ligands highlighted the crucial residues to be targeted for the inhibition of the helicase activity of DDX3. The obtained results confirm the reliability of the constructed DDX3/RNA model and the proposed computational strategy for investigating novel DDX3 inhibitors.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Targeting cellular cofactors instead of viral enzymes represents a new strategy to combat infectious diseases, which should help to overcome the problem of viral resistance. Recently, it has been revealed that the cellular ATPase/RNA helicase X-linked DEAD-box polypeptide 3 (DDX3) is an essential host factor for the replication of several viruses such as HIV, HCV, JEV, Dengue, and West Nile. Accordingly, a drug targeting DDX3 could theoretically inhibit all viruses that are dependent on this host factor. Herein, for the first time, a model of hDDX3 in its closed conformation, which binds the viral RNA was developed by using the homology module of Prime through the Maestro interface of Schrodinger. Next, a structure-based virtual screening protocol was applied to identify DDX3 small molecule inhibitors targeting the RNA binding pocket. As a result, an impressive hit rate of 40% was obtained with the identification of 10 active compounds out of the 25 tested small molecules. The best poses of the active ligands highlighted the crucial residues to be targeted for the inhibition of the helicase activity of DDX3. The obtained results confirm the reliability of the constructed DDX3/RNA model and the proposed computational strategy for investigating novel DDX3 inhibitors. |
Bavagnoli L; Cucuzza S; Campanini G; Rovida F; Paolucci S; Baldanti F; Maga G The novel influenza A virus protein PA-X and its naturally deleted variant show different enzymatic properties in comparison to the viral endonuclease PA. Journal Article In: Nucleic Acids Research, vol. 43, no 19, pp. 9405-9417, 2015. @article{%a1:%Y_414,
title = {The novel influenza A virus protein PA-X and its naturally deleted variant show different enzymatic properties in comparison to the viral endonuclease PA.},
author = {Bavagnoli L and Cucuzza S and Campanini G and Rovida F and Paolucci S and Baldanti F and Maga G},
url = {https://academic.oup.com/nar/article/43/19/9405/2528196},
doi = {10.1093/nar/gkv926},
year = {2015},
date = {2015-10-15},
journal = {Nucleic Acids Research},
volume = {43},
number = {19},
pages = {9405-9417},
abstract = {The PA protein of Influenza A virus (IAV) encoded by segment 3 acts as a specialized RNA endonuclease in the transcription of the viral genome. The same genomic segment encodes for a second shorter protein, termed PA-X, with the first 191 N-terminal aminoacids (aa) identical to PA, but with a completely different C-ter domain of 61 aa, due to a ribosomal frameshifting. In addition, it has been shown that several IAV isolates encode for a naturally truncated PA-X variant, PAXΔC20, missing the last 20 aa. The biochemical properties of PA-X and PAXΔC20 have been poorly investigated so far. Here, we have carried out an enzymatic characterization of PA-X and its naturally deleted form, in comparison with PA from the human IAV strain A/WSN/33 (H1N1). Our results showed, to the best of our knowledge for the first time, that PA-X possesses an endonucleolytic activity. Both PA and PA-X preferentially cut single stranded RNA regions, but with some differences. In addition, we showed that PAXΔC20 has severely reduced nuclease activity. These results point to a previously undetected role of the last C-ter 20 aa for the catalytic activity of PA-X and support distinct roles for these proteins in the viral life cycle. The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
The PA protein of Influenza A virus (IAV) encoded by segment 3 acts as a specialized RNA endonuclease in the transcription of the viral genome. The same genomic segment encodes for a second shorter protein, termed PA-X, with the first 191 N-terminal aminoacids (aa) identical to PA, but with a completely different C-ter domain of 61 aa, due to a ribosomal frameshifting. In addition, it has been shown that several IAV isolates encode for a naturally truncated PA-X variant, PAXΔC20, missing the last 20 aa. The biochemical properties of PA-X and PAXΔC20 have been poorly investigated so far. Here, we have carried out an enzymatic characterization of PA-X and its naturally deleted form, in comparison with PA from the human IAV strain A/WSN/33 (H1N1). Our results showed, to the best of our knowledge for the first time, that PA-X possesses an endonucleolytic activity. Both PA and PA-X preferentially cut single stranded RNA regions, but with some differences. In addition, we showed that PAXΔC20 has severely reduced nuclease activity. These results point to a previously undetected role of the last C-ter 20 aa for the catalytic activity of PA-X and support distinct roles for these proteins in the viral life cycle. The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research. |
Tintori C; La Sala G; Vignaroli G; Botta L; Fallacara AL; Falchi F; Radi M; Zamperini C; Dreassi E; Dello Iacono L; Orioli D; Biamonti G; Garbelli M; Lossani A; Gasparrini F; Tuccinardi T; Laurenzana I; Angelucci A; Maga G; Schenone S; Brullo C; Musumeci F; Desogus A; Crespan E; Botta M Studies on the ATP Binding Site of Fyn Kinase for the Identification of New Inhibitors and Their Evaluation as Potential Agents against Tauopathies and Tumors. Journal Article In: Journal of Medicinal Chemistry, vol. 58, no 11, pp. 4590-4609, 2015. @article{%a1:%Y_419,
title = {Studies on the ATP Binding Site of Fyn Kinase for the Identification of New Inhibitors and Their Evaluation as Potential Agents against Tauopathies and Tumors.},
author = {Tintori C and La Sala G and Vignaroli G and Botta L and Fallacara AL and Falchi F and Radi M and Zamperini C and Dreassi E and Dello Iacono L and Orioli D and Biamonti G and Garbelli M and Lossani A and Gasparrini F and Tuccinardi T and Laurenzana I and Angelucci A and Maga G and Schenone S and Brullo C and Musumeci F and Desogus A and Crespan E and Botta M},
url = {https://pubs.acs.org/doi/10.1021/acs.jmedchem.5b00140},
doi = {10.1021/acs.jmedchem.5b00140},
year = {2015},
date = {2015-06-11},
journal = {Journal of Medicinal Chemistry},
volume = {58},
number = {11},
pages = {4590-4609},
abstract = {Fyn is a member of the Src-family of nonreceptor protein-tyrosine kinases. Its abnormal activity has been shown to be related to various human cancers as well as to severe pathologies, such as Alzheimer's and Parkinson's diseases. Herein, a structure-based drug design protocol was employed aimed at identifying novel Fyn inhibitors. Two hits from commercial sources (1, 2) were found active against Fyn with K(i) of about 2 μM, while derivative 4a, derived from our internal library, showed a K(i) of 0.9 μM. A hit-to-lead optimization effort was then initiated on derivative 4a to improve its potency. Slightly modifications rapidly determine an increase in the binding affinity, with the best inhibitors 4c and 4d having K(i)s of 70 and 95 nM, respectively. Both compounds were found able to inhibit the phosphorylation of the protein Tau in an Alzheimer's model cell line and showed antiproliferative activities against different cancer cell lines.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Fyn is a member of the Src-family of nonreceptor protein-tyrosine kinases. Its abnormal activity has been shown to be related to various human cancers as well as to severe pathologies, such as Alzheimer's and Parkinson's diseases. Herein, a structure-based drug design protocol was employed aimed at identifying novel Fyn inhibitors. Two hits from commercial sources (1, 2) were found active against Fyn with K(i) of about 2 μM, while derivative 4a, derived from our internal library, showed a K(i) of 0.9 μM. A hit-to-lead optimization effort was then initiated on derivative 4a to improve its potency. Slightly modifications rapidly determine an increase in the binding affinity, with the best inhibitors 4c and 4d having K(i)s of 70 and 95 nM, respectively. Both compounds were found able to inhibit the phosphorylation of the protein Tau in an Alzheimer's model cell line and showed antiproliferative activities against different cancer cell lines. |
Crespan E; Hübscher U; Maga G Expansion of CAG triplet repeats by human DNA polymerases λ and β in vitro, is regulated by flap endonuclease 1 and DNA ligase 1. Journal Article In: DNA Repair, vol. 29, pp. 101-111, 2015. @article{%a1:%Y_361,
title = {Expansion of CAG triplet repeats by human DNA polymerases λ and β in vitro, is regulated by flap endonuclease 1 and DNA ligase 1.},
author = {Crespan E and Hübscher U and Maga G},
url = {https://www.sciencedirect.com/science/article/pii/S1568786415000178?via%3Dihub},
doi = {10.1016/j.dnarep.2015.01.005},
year = {2015},
date = {2015-05-29},
journal = {DNA Repair},
volume = {29},
pages = {101-111},
abstract = {Huntington's disease (HD) is a neurological genetic disorder caused by the expansion of the CAG trinucleotide repeats (TNR) in the N-terminal region of coding sequence of the Huntingtin's (HTT) gene. This results in the addition of a poly-glutamine tract within the Huntingtin protein, resulting in its pathological form. The mechanism by which TRN expansion takes place is not yet fully understood. We have recently shown that DNA polymerase (Pol) beta can promote the microhomology-mediated end joining andtriplet expansion of a substrate mimicking a double strand break in the TNR region of the HTT gene. Here we show that TNRexpansion is dependent on the structure of the DNA substrate, as well as on the two essential Pol beta co-factors: flap endonuclease1 (Fen1) and DNA ligase 1 (Lig1). We found that Fen1 significantly stimulated TNR expansion by Pol beta, but not by the related enzyme Pol lambda, and subsequent ligation of the DNA products by Lig1. Interestingly, the deletion of N-terminal domains of Pol lambda, resulted in an enzyme which displayed properties more similar to Pol beta, suggesting a possible evolutionary mechanism. These results may suggest a novel mechanism for somatic TNR expansion in HD.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
Huntington's disease (HD) is a neurological genetic disorder caused by the expansion of the CAG trinucleotide repeats (TNR) in the N-terminal region of coding sequence of the Huntingtin's (HTT) gene. This results in the addition of a poly-glutamine tract within the Huntingtin protein, resulting in its pathological form. The mechanism by which TRN expansion takes place is not yet fully understood. We have recently shown that DNA polymerase (Pol) beta can promote the microhomology-mediated end joining andtriplet expansion of a substrate mimicking a double strand break in the TNR region of the HTT gene. Here we show that TNRexpansion is dependent on the structure of the DNA substrate, as well as on the two essential Pol beta co-factors: flap endonuclease1 (Fen1) and DNA ligase 1 (Lig1). We found that Fen1 significantly stimulated TNR expansion by Pol beta, but not by the related enzyme Pol lambda, and subsequent ligation of the DNA products by Lig1. Interestingly, the deletion of N-terminal domains of Pol lambda, resulted in an enzyme which displayed properties more similar to Pol beta, suggesting a possible evolutionary mechanism. These results may suggest a novel mechanism for somatic TNR expansion in HD. |
Spallarossa A; Caneva C; Caviglia M; Alfei S; Butini S; Campiani G; Gemma S; Brindisi M; Zisterer DM; Bright SA; Williams CD; Crespan E; Maga G; Sanna G; Delogu I; Collu G; Loddo R Unconventional Knoevenagel-type indoles: Synthesis and cell-based studies for the identification of pro-apoptotic agents. Journal Article In: European Journal of Medicinal Chemistry, vol. 102, pp. 648-660, 2015. @article{%a1:%Y_418,
title = {Unconventional Knoevenagel-type indoles: Synthesis and cell-based studies for the identification of pro-apoptotic agents.},
author = {Spallarossa A and Caneva C and Caviglia M and Alfei S and Butini S and Campiani G and Gemma S and Brindisi M and Zisterer DM and Bright SA and Williams CD and Crespan E and Maga G and Sanna G and Delogu I and Collu G and Loddo R},
url = {https://www.sciencedirect.com/science/article/pii/S0223523415301938?via%3Dihub},
doi = {10.1016/j.ejmech.2015.08.009},
year = {2015},
date = {2015-03-12},
urldate = {2017-03-03},
journal = {European Journal of Medicinal Chemistry},
volume = {102},
pages = {648-660},
abstract = {A new series of indole-based analogues were recently identified as potential anticancer agents. The Knoevenagel-type indoles herein presented were prepared via a one-pot condensation of iminium salts with active methylene reagents and were isolated as single geometric isomers. Biological evaluation in different cell-based assays revealed an antiproliferative activity for some analogues already in the nanomolar range against leukaemia, breast and renal cancer cell lines. To explain these effects, the most promising analogues of the series were engaged in further cell-based studies. Compounds 5e, l, p and 6a, b highlighted a pro-apoptotic potential being able to induce apoptosis in HL60, K562 and MCF-7 cell lines in a dose and time-dependent manner. The ability of these compounds to arrest cell cycle at the G2/M phase inspired the immunofluorescence studies which allowed us to identify tubulin as a potential target for compounds 5l and 6b. Copyright 2015 Elsevier Masson SAS. All rights reserved.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
A new series of indole-based analogues were recently identified as potential anticancer agents. The Knoevenagel-type indoles herein presented were prepared via a one-pot condensation of iminium salts with active methylene reagents and were isolated as single geometric isomers. Biological evaluation in different cell-based assays revealed an antiproliferative activity for some analogues already in the nanomolar range against leukaemia, breast and renal cancer cell lines. To explain these effects, the most promising analogues of the series were engaged in further cell-based studies. Compounds 5e, l, p and 6a, b highlighted a pro-apoptotic potential being able to induce apoptosis in HL60, K562 and MCF-7 cell lines in a dose and time-dependent manner. The ability of these compounds to arrest cell cycle at the G2/M phase inspired the immunofluorescence studies which allowed us to identify tubulin as a potential target for compounds 5l and 6b. Copyright 2015 Elsevier Masson SAS. All rights reserved. |
Tintori C; Fallacara AL; Radi M; Zamperini C; Dreassi E; Crespan E; Maga G; Schenone S; Musumeci F; Brullo C; Richters A; Gasparrini F; Angelucci A; Festuccia C; Delle Monache S; Rauh D; Botta M Combining X-ray Crystallography and Molecular Modeling toward the Optimization of Pyrazolo[3,4-d]pyrimidines as Potent c-Src Inhibitors Active in Vivo against Neuroblastoma. Journal Article In: Journal of Medicinal Chemistry, vol. 58, no 1, pp. 347-361, 2015. @article{%a1:%Y_348,
title = {Combining X-ray Crystallography and Molecular Modeling toward the Optimization of Pyrazolo[3,4-d]pyrimidines as Potent c-Src Inhibitors Active in Vivo against Neuroblastoma.},
author = {Tintori C and Fallacara AL and Radi M and Zamperini C and Dreassi E and Crespan E and Maga G and Schenone S and Musumeci F and Brullo C and Richters A and Gasparrini F and Angelucci A and Festuccia C and {Delle Monache S} and Rauh D and Botta M},
url = {https://pubs.acs.org/doi/10.1021/jm5013159},
doi = {10.1021/jm5013159},
year = {2015},
date = {2015-02-12},
journal = {Journal of Medicinal Chemistry},
volume = {58},
number = {1},
pages = {347-361},
abstract = {c-Src is a tyrosine kinase belonging to the Src-family kinases. It is overexpressed and/or hyperactivated in a variety of cancer cells, thus its inhibition has been predicted to have therapeutic effects in solid tumors. Recently, the pyrazolo[3,4-d]pyrimidine 3 was reported as a dual c-Src/Abl inhibitor. Herein we describe a multidisciplinary drug discovery approach for the optimization of the lead 3 against c-Src. Starting from the X-ray crystal structure of c-Src in complex with 3, Monte Carlo free energy perturbation calculations were applied to guide the design of c-Src inhibitors with improved activities. As a result, the introduction of a meta hydroxyl group on the C4 anilino ring was computed to be particularly favorable. The potency of the synthesized inhibitors was increased with respect to the starting lead 3. The best identified compounds were also found active in the inhibition of neuroblastoma cell proliferation. Furthermore, compound 29 also showed in vivo activity in xenograft model using SH-SY5Y cells.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
c-Src is a tyrosine kinase belonging to the Src-family kinases. It is overexpressed and/or hyperactivated in a variety of cancer cells, thus its inhibition has been predicted to have therapeutic effects in solid tumors. Recently, the pyrazolo[3,4-d]pyrimidine 3 was reported as a dual c-Src/Abl inhibitor. Herein we describe a multidisciplinary drug discovery approach for the optimization of the lead 3 against c-Src. Starting from the X-ray crystal structure of c-Src in complex with 3, Monte Carlo free energy perturbation calculations were applied to guide the design of c-Src inhibitors with improved activities. As a result, the introduction of a meta hydroxyl group on the C4 anilino ring was computed to be particularly favorable. The potency of the synthesized inhibitors was increased with respect to the starting lead 3. The best identified compounds were also found active in the inhibition of neuroblastoma cell proliferation. Furthermore, compound 29 also showed in vivo activity in xenograft model using SH-SY5Y cells. |
Vincetti P; Caporuscio F; Kaptein S; Gioiello A; Mancino V; Suzuki Y; Yamamoto N; Crespan E; Lossani A; Maga G; Rastelli G; Castagnolo D; Neyts J; Leyssen P; Costantino G; Radi M Discovery of Multitarget Antivirals Acting on Both the Dengue Virus NS5-NS3 Interaction and the Host Src/Fyn Kinases. Journal Article In: Journal of Medicinal Chemistry, vol. 58, no 12, 2015. @article{%a1:%Y_353,
title = {Discovery of Multitarget Antivirals Acting on Both the Dengue Virus NS5-NS3 Interaction and the Host Src/Fyn Kinases.},
author = {Vincetti P and Caporuscio F and Kaptein S and Gioiello A and Mancino V and Suzuki Y and Yamamoto N and Crespan E and Lossani A and Maga G and Rastelli G and Castagnolo D and Neyts J and Leyssen P and Costantino G and Radi M},
url = {https://pubs.acs.org/doi/10.1021/acs.jmedchem.5b00108},
doi = {10.1021/acs.jmedchem.5b00108},
year = {2015},
date = {2015-02-12},
journal = {Journal of Medicinal Chemistry},
volume = {58},
number = {12},
abstract = {This study describes the discovery of novel dengue virus inhibitors targeting both a crucial viral protein-protein interaction and an essential host cell factor as a strategy to reduce the emergence of drug resistance. Starting from known c-Src inhibitors, a virtual screening was performed to identify molecules able to interact with a recently discovered allosteric pocket on the dengue virus NS5 polymerase. The selection of cheap-to-produce scaffolds and the exploration of the biologically relevant chemical space around them suggested promising candidates for chemical synthesis. A series of purines emerged as the most interesting candidates able to inhibit virus replication at low micromolar concentrations with no significant toxicity to the host cell. Among the identified antivirals, compound 16i proved to be 10 times more potent than ribavirin, showed a better selectivity index and represents the first-in-class DENV-NS5 allosteric inhibitor able to target both the virus NS5-NS3 interaction and the host kinases c-Src/Fyn.},
keywords = {},
pubstate = {published},
tppubtype = {article}
}
This study describes the discovery of novel dengue virus inhibitors targeting both a crucial viral protein-protein interaction and an essential host cell factor as a strategy to reduce the emergence of drug resistance. Starting from known c-Src inhibitors, a virtual screening was performed to identify molecules able to interact with a recently discovered allosteric pocket on the dengue virus NS5 polymerase. The selection of cheap-to-produce scaffolds and the exploration of the biologically relevant chemical space around them suggested promising candidates for chemical synthesis. A series of purines emerged as the most interesting candidates able to inhibit virus replication at low micromolar concentrations with no significant toxicity to the host cell. Among the identified antivirals, compound 16i proved to be 10 times more potent than ribavirin, showed a better selectivity index and represents the first-in-class DENV-NS5 allosteric inhibitor able to target both the virus NS5-NS3 interaction and the host kinases c-Src/Fyn. |
2014
|
Maga G AIDS: la verità negata Book Il Pensiero Scientifico Editore, 2014, ISBN: 978-8849004809, (Vincitore del primo premio categoria saggi del Premio Letterario Nazionale Fanz Kafka - 2014). @book{CNRPRODOTTI303054,
title = {AIDS: la verità negata},
author = {Maga G},
url = {http://www.pensiero.it/ecomm/pc/viewPrd.asp?idproduct=651},
isbn = {978-8849004809},
year = {2014},
date = {2014-01-01},
publisher = {Il Pensiero Scientifico Editore},
series = {Informa},
abstract = {Ancora oggi esiste una corrente di opinione, tutt’altro che secondaria e sempre vitale, che afferma che l’AIDS non esiste e che l’HIV è un innocuo parassita. Ma negare l’AIDS significa sottovalutare la pericolosità dell’infezione da HIV e indurre i pazienti a rifiutare terapie in grado di salvare loro la vita. Quando l’opinione errata di alcuni compromette la salute e la sicurezza di altri, è necessario correggerla. Giovanni Maga dimostra, con un linguaggio comprensibile a tutti e attraverso la narrazione appassionata di tante storie di pazienti, medici e ricercatori, che respingere la relazione tra HIV e AIDS è un errore dalle drammatiche conseguenze. Documentando, inoltre, gli straordinari progressi fatti nel combattere questa malattia, il libro vuole essere un invito all’ottimismo e alla speranza. La difficoltà nell’accettare l’AIDS e, di conseguenza, la disponibilità a credere che non esista nascono spesso dalla disperazione di chi vive la sieropositività come una condanna senza appello. Ma la scienza arriverà inevitabilmente a sconfiggere l’HIV e questo libro ci spiega come e perché.},
note = {Vincitore del primo premio categoria saggi del Premio Letterario Nazionale Fanz Kafka - 2014},
keywords = {},
pubstate = {published},
tppubtype = {book}
}
Ancora oggi esiste una corrente di opinione, tutt’altro che secondaria e sempre vitale, che afferma che l’AIDS non esiste e che l’HIV è un innocuo parassita. Ma negare l’AIDS significa sottovalutare la pericolosità dell’infezione da HIV e indurre i pazienti a rifiutare terapie in grado di salvare loro la vita. Quando l’opinione errata di alcuni compromette la salute e la sicurezza di altri, è necessario correggerla. Giovanni Maga dimostra, con un linguaggio comprensibile a tutti e attraverso la narrazione appassionata di tante storie di pazienti, medici e ricercatori, che respingere la relazione tra HIV e AIDS è un errore dalle drammatiche conseguenze. Documentando, inoltre, gli straordinari progressi fatti nel combattere questa malattia, il libro vuole essere un invito all’ottimismo e alla speranza. La difficoltà nell’accettare l’AIDS e, di conseguenza, la disponibilità a credere che non esista nascono spesso dalla disperazione di chi vive la sieropositività come una condanna senza appello. Ma la scienza arriverà inevitabilmente a sconfiggere l’HIV e questo libro ci spiega come e perché. |