From 3D chromatin structure to gene regulation.
A recent study coordinated by Francesco Ferrari – Institute of Molecular Genetics “Luigi Luca Cavalli-Sforza” of the National Research Council in Pavia (IGM-CNR) and IFOM, the FIRC Institute of Molecular Oncology – demonstrated how the physical three-dimensional (3D) conformation of DNA inside the cell nucleus can be leveraged to reconstruct the network of regulatory interactions controlling genes activity.
The 3D organization of DNA and its associated proteins (chromatin) inside the cell nucleus is regulated by biophysical principles and biochemical processes to comply with structural and functional needs. In particular, several genetic regulatory elements (enhancers and promoters) need to physically interact with each other for proper control of gene activity. As these regulatory elements are sometimes positioned at a long distance along the linear sequence of the DNA, a specific 3D configuration of chromatin is required to bring them in close physical proximity.
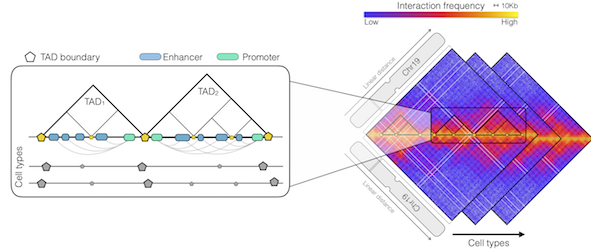
This study shows that a rigorous statistical framework incorporating data on chromatin 3D is effective in improving the reconstruction of these crucial distal regulatory interactions. This work will provide the basis to improve the characterization of genetic mutations or functional alterations of DNA regulatory elements in cancer and genetic diseases.
This study has been supported by AIRC Foundation for cancer research, which also funded a fellowship for a young researcher, Dr. Elisa Salviato, first author of the article.
