Phosphorylation-mediate control of RNA editing by AKT in tumorigenesis and cancer progression
Tumorigenesis and cancer progression involve a series of cellular pathways that control cell growth and proliferation. Any protein associated with these pathways may be a potential oncogene or tumor suppressor gene. Current molecular factors involved in the initiation of cancer include genomic instability and somatic mutations, but also epigenetic alterations and modification of the transcriptome are emerging as a significant force during the transition from normal to malignant cells. Editing of adenosine to inosine (A-to-I) in double stranded RNA catalyzed by adenosine deaminase acting on RNA (ADARs) is a dynamic modification that can perturb the transcriptome: the interpretation of I as a guanosine (G) can lead to non-synonymous codon changes, alternative splicing and perturbation of miRNA metabolism (Xu LD et al., 2018). Uncontrolled nucleotide changes can result in susceptibility of cells to developmental defects and potential carcinogenicity. This aspect of RNA editing and how it is regulated, is still largely unexplored. Adenosine deaminase acting on RNA (ADARs) catalyze the conversion of adenosine (A) to inosine (I) in double-stranded RNA. Inosine exhibits similar properties as guanosine. RNA editing can result in post-transcriptional changes in codons, diversifying transcripts and giving rise to an enormous variety in the cell proteome, but also in cellular development defects and in potential carcinogenic susceptibility. RNA editing is a fundamental modification event that controls nucleotide changes bypassing the genomic information. Mammalian cells express three ADAR proteins, ADAR1, ADAR2 and ADAR3, the last shown to be inactive for RNA editing.
Abnormal ADARs editing is present in a variety of cancers (hepatocellular carcinoma, chronic myeloid leukemia, multiple myeloma, glioblastoma). ADAR1 is typically upregulated and supports growth and cell survival by editing mRNAs and miRNAs involved in cancer promotion, although it sometimes serves in tumor suppression, while ADAR2 editing inhibits tumor growth.
Recently, we defined a novel role for the AKT family kinases in the regulation of “RNA editing” (Piazzi et al., 2020; Bavelloni et al., 2019): active AKT kinases were found to phosphorylate the double-stranded RNA adenosine deaminases (ADAR1 and ADAR2) on a conserved threonine within the catalytic domain as well as four additional sites (3 in ADAR1 and 1 in ADAR2) (Fig.1). Mimicking phosphorylation in the catalytic domain resulted in a differential reduction in editase activity toward known ADAR substrates. Thus, AKT-mediated phosphorylation of ADAR-1/-2 may represent a direct link tying aberrant PI3K-AKT-mTOR pathway activation to tumor-associated alterations in splicing. In order to determine whether the status of AKT activation is directly associated to alternative splicing abnormalities and whether targeting the AKT-ADAR axis is a viable option to enhance the therapeutic response in cancers, our Lab is trying to answer the following question:
- What effect the AKT activation status has on ADAR1 and ADAR2 isoform expression?
- What effect AKT-dependent phosphorylation of the individual ADAR1 and ADAR2 isoforms has on global editing/splicing events in the cell?
- What influence AKT-dependent ADAR1 and ADAR2 phosphorylation has on tumorigenesis?
- What influence AKT-dependent ADAR1 and ADAR2 phosphorylation has on therapeutic response?
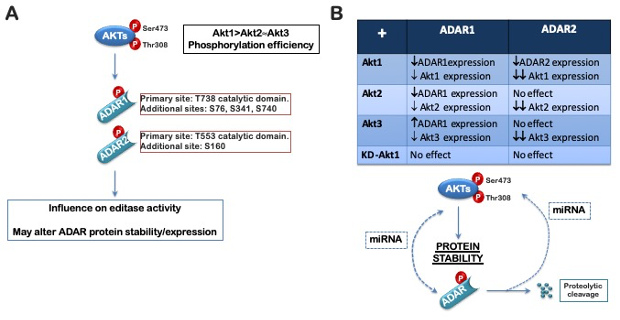 |
Figure 1. Schematic diagram of AKT-dependent regulation of ADAR1 and ADAR2. (A) All three AKT isoforms phosphorylate both ADAR1p110 and ADAR2. Four sites were found to be phosphorylated by AKT1 in ADAR1p110 (S76, S341, T738, S740), with the major phosphorylation
site being T738; two sites were found to be phosphorylated by AKT1 in ADAR2 (S160 and T553), with the major phosphorylated site being T553. (B) Active AKT and the ADARs may co-regulate one another. The exact mechanism has not been defined, but AKT might influence ADAR1p110 and ADAR2 protein stability or ADAR. |
Bibliography:
Xu LD and Ohman M. ADAR1 editing and its role in cancer. (2018). Genes (Basel). 25;10(1):12.
Piazzi M, Bavelloni A, Gallo A, Blalock WL. AKT-Dependent Phosphorylation of ADAR1p110 and ADAR2 Represents a New and Important Link Between Cell Signaling and RNA Editing. (2020). DNA Cell Biol. 39(3):343-348.
Bavelloni A, Focaccia E, Piazzi M, Raffini M, Cesarini V, Tomaselli S, et al. AKT-dependent phosphorylation of the adenosine deaminases ADAR-1 and -2 inhibits deaminase activity. (2019). FASEB J. 33:9044–61.
